Below
a collection of programs & modules developed by Scott Davis in either
R, Java or MATLAB for the analysis of microarray gene expression.
They are all designed to work with the same input file format:
a
GenePattern Expression Dataset
(*.gct). All of these programs can be installed
as
GenePattern
modules, though some of them can also be run as stand-alone
Java applications (as indicated by the Java logo).

GENEPATTERN Module |
In GenePattern, under the
Modules & Pipelines menu, select "Install
from zip" and specify the corresponding ZIP
file.
NOTE:
Custom modules can only be installed on local installations
of GenePattern. If you are logging into the Broad's
server, these modules cannot be installed. |

JAVA Application |
Any programs with the
Java Webstart launch button can
be started directly from this page (you may get one
ore more security prompts, just click accept/allow).
After running a program for the first time, a shortcut
will appear on your desktop as well as in your applications
(Start) menu under the "CBDM Bioinformatics
Tools" folder. Any program updates will automatically
be downloaded whenever you run them from your computer.
NOTE:
It is recommended to install Java 64bit whenever possible,
and ensure that your browser is calling the 64bit executable
whenever launching *.jnlp files (look under MIME/Application
settings in browser options). |
| Example Data |
|
|
Bioinformatics Software |
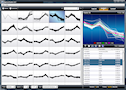
ExpressCluster
|
Interactive K-means clustering of genes based on their expression
profiles across a variable number of conditions. User-friendly
GUI allows researchers to visualize results instantly in
order to generate complex gene lists/signatures, heatmaps,
and new expression datasets based on computed clusters.
Export results in PNG, PDF, SVG and EPS.
Version
1.3 has critical fixes to the search feature, as well as
additional search options by right-clicking a probe in the
cluster data table. |
1.3
|
2GB RAM
1GB RAM
 ExpressCluster_v1.3.zip ExpressCluster_v1.3.zip
 ExpressCluster_v1.3.pdf ExpressCluster_v1.3.pdf |
Population PCA
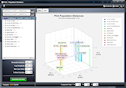 |
PCA-based 3D plotting of samples/populations. Resulting
plot can be rotated in real time, and populations can be
reordered by drag & drop in the legend. Drop lines, populations
labels, and color customizations are also available.
NOTE: The MATLAB version requires that
the Matlab Compiler Runtime (included in zip) be installed,
and currently will only run on Windows x64 systems (Vista
or Win7). The only advantage to using this version is the
ability to export plots in vector based formats (EPS, AI,
Figure) for further modifications to the plot. However,
this version does not have the same interactive capabilities
as the Java version. |
2.0
|
1GB RAM
 PopulationPCA-Matlab.zip PopulationPCA-Matlab.zip |
GCT Extractor
 |
Graphical tool to create subsets from a master GCT (along
with the accompanying CLS file). Users can also reorder
popuations. |
1.0
|

|
ExpressMatrix
 |
Quickly view expression/expression plots and correlations
for multiple samples at once.
NOTE:
This program is quite old (3+ years) and is in serious need
of an upgrade. I will try to post that in the next few months.
|
1.0
|
 ExpressMatrix.zip ExpressMatrix.zip
 ExpressMatrix.pdf ExpressMatrix.pdf |
δ-Score
 |
Computes a score for differential expression between two
populations. The score is essentially a noise-adjusted measure
of fold-change (somewhat similar to ANOVA). |
1.0 |
 DeltaScore.zip DeltaScore.zip
|
|
ProbesetCorrelationCluster |
Generates heatmaps of a hierarchically clustered correlation
matrix for a subset of probes. |
1.0 |
ProbesetCorrCluster.zip |
|
ExtractProbes |
Extract a subset a probes from a GCT based on a list of
either probeIDs or gene symbols (the latter assumes that
the input GCT contains gene symbols as its annotation).
Output is a new GCT containing the matched probes. |
1.2 |
 ExtractProbes.zip ExtractProbes.zip |
|
ClassMeans |
Generates a class means GCT (and CLS) from one containing
replicate samples. |
1.0 |
 ClassMeans.zip ClassMeans.zip |
Department of Microbiology & Immunobiology • Harvard Medical
School • 77 Avenue Louis Pasteur • Boston, MA 02115
|




